Research Projects
The central goal of the project area A is to gain a global view of adaptive processes in respiratory pathogens. Proteins, especially enzymes, are the functional key players in infection, and proteome and metabolome dynamics reflect the adaptation of microorganisms to their different niches in the respiratory tract. Strategies and techniques for OMICs analysis developed for S. aureus in CRC-TRR 34 “Pathophysiology of Staphylococci in the Post-Genomic Era” projects can now be transferred to investigate the transcriptome, proteome and metabolome signatures of S. pneumoniae and B. pseudomallei under various in vitro stress exposures or in vivo conditions.
During the infection process a pathogen faces several defence strategies. The pathogen can tolerate some of these onslaughts due to specific stress response mechanisms. In contrast to other pathogens such as S. aureus, almost nothing is known about the basic stress physiology and specific stress responses in B. pseudomallei or S. pneumoniae. Only a very few investigations into such stress responses in B. pseudomallei and S. pneumoniae at metabolome and proteome level are available. In this project we will focus on in vitro analyses of infection-related stress responses of S. pneumoniae and B. pseudomallei at the metabolome and proteome level to better understand the infection process and defence mechanisms of these organisms. We will therefore analyse different physiological conditions such as nutrient starvation, iron limitation and anaerobiosis.
Principal Investigators
Prof. Dr. rer. nat. Dörte Becher
Prof. Dr. rer. nat. Michael Lalk
Thesis Topics
1. Juliane Hoyer
"Investigation of infection-relevant stresses in Streptococcus pneumoniae"
2. Anne Leonard
"Exploring metabolic adaptation of Streptococcus pneumoniae to environmental stresses"
Bacterial adaptation reactions to environmental insults have been extensively studied under laboratory conditions in vitro at a genome-wide level for model bacteria including Bacillus subtilis and S. aureus. However, for pathogenic bacteria these studies only incompletely capture infection settings. To analyze the interplay between pathogens and hosts at a global scale OMICs studies have been extended to cell culture and animal models. While the host side can be analysed well using these approaches, studying the pathogen is far more complicated due to limited availability of material. Proteomic approaches provide additional information to transcriptomics but are particularly challenging and scarce due to lower sensitivity and high background of host proteins. Relying on expertise in proteome/transcriptome studies with Staphylococcus aureus, this project focuses on the in vivo adaptation of B. pseudomallei and S. pneumoniae using a combination of transcriptomic and proteomic approaches.
Principal Investigators
Prof. Dr. rer. nat. Dörte Becher
Prof. Dr. rer. nat. Uwe Völker
1. Claudia Hirschfeld
"Analysis of the phosphoproteome of Streptococcus pneumoniae"
2. Anna Nagel
"Comparative proteome/transcriptome analyses of the adaptation of Staphylococcus aureus and Burkholderia pseudomallei to host cells"
Streptococcus pneumoniae is an emerging pathogen with an intracellular lifestyle that has to adapt to changing environmental conditions during infection processes. These adaptations are closely connected to the host environmental metabolite composition. Recent studies have shown that some changes in the pathogen’s metabolism during an infection are caused by the host response, and cannot be detected in chemically defined media in vitro. Thus, the nutrient status for the pathogen inside the host cell is determined by the host cell’s metabolism and cannot serve as growth conditions for non-adapted bacteria. Therefore, both the invading pathogen and defending host cell likely undergo metabolic adaption. Pathogens able to replicate inside the host cell have already been used in infection studies to investigate how they can use host-derived carbon sources. A more comprehensive study of how pathogens like S. pneumoniae alter the metabolome will provide essential data for systems biology approaches. In this project we will focus on investigating the metabolic impact of pneumococcal-host interactions in in vitro and cell culture experiments.
Principal Investigators
Prof. Dr. rer. nat. Michael Lalk
Thesis Topics
1. Daniel Schultz
"Metabolome analysis in bacto-viral infection studies"
2. Stefanie Schneider
"Host-pathogen interactions: metabolic adaptation mechanisms"
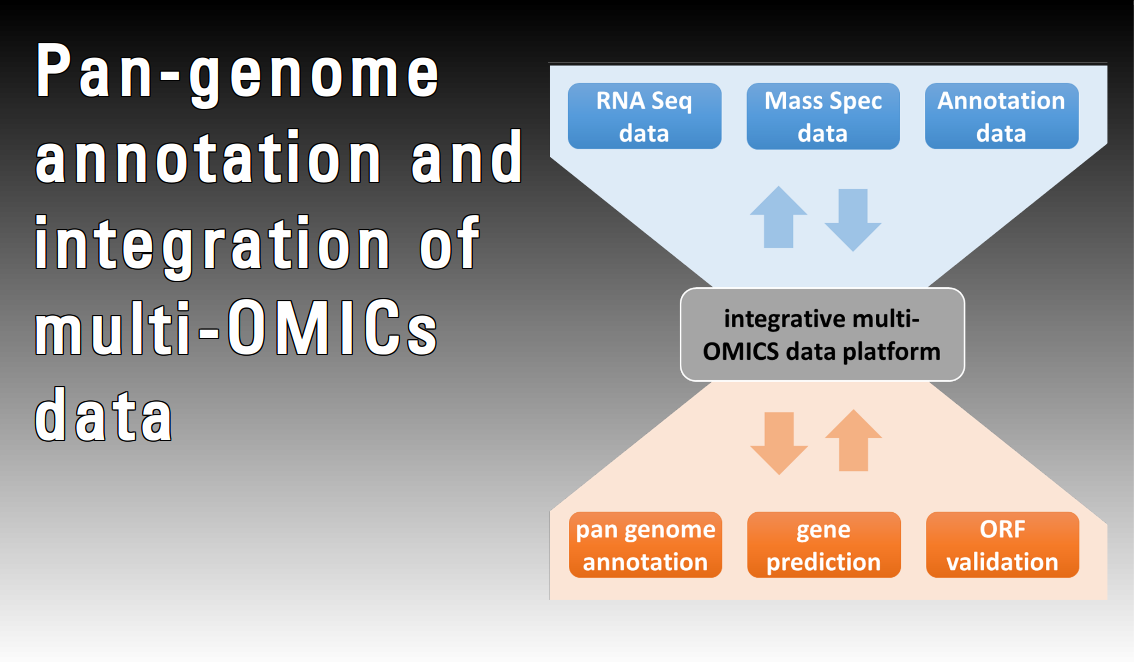
Bacterial genomes are still typically annotated one genome at a time. However, a recent study comparing individually annotated genomes from multiple isolates of the same species, pan-genomes, found that these gene structures were very often inconsistent with each other. Considering the increasing availability of pan-genomes it is suggested that de-novo gene finders should integrate information from a multiple pan-genome alignment. Furthermore, gene expression analyses – especially RNA-Seq – can contribute substantially to more precise annotation, particularly for small non-coding RNAs. Nevertheless, only a few studies are taking advantage of an integrative approach combining results from different OMICs techniques, and thus providing a look beyond the “DNA/RNA world”. In this project methods for improved prokaryotic pan-genome annotation will be developed with an emphasis on new approaches made feasible by 1) a rapidly increasing number of genomes from the same bacterial species, 2) high-throughput proteomics and 3) RNA-Seq.
Principal Investigators
Prof. Dr. rer. nat. Mario Stanke
Thesis Topics
Lars Romoth
"Combining proteogenomics, transcriptomics and comparative genomics for bacterial pan-genome annotation"

Cell cultures constitute a simplified model to study host-pathogen-interactions at the molecular level. They provide sufficient material for the simultaneous analysis of host and pathogen under highly standardized conditions. Unfortunately, these systems do not really reflect the complex situation in animal models or the human body, where multiple cell types simultaneously contribute to interactions with bacterial pathogens. Still, cell culture models have their value since in in vivo models deciphering molecular mechanisms is complicated by the presence of numerous immune cell populations and multiple interacting regulatory pathways. In an attempt to provide a link between both extremes we want to extend current in vitro cell culture studies into a three-component system where one can investigate the interplay between two different host cell types interacting with bacterial pathogens.
Principal Investigators
Prof. Dr. rer. nat. Uwe Völker
Thesis Topics
1. Laura M. Palma Medina
"Characterization of the modification of Staphylococcus aureus-epithelial mucosa interaction by neutrophils in a complex three partner in vitro cell culture model"
2. Solomon Mekonnen
"Interaction of S. aureus MRSA with its host: A multi-omics approach"
Bacterial virulence is determined by proficient adherence, colonization, invasion, adaptive fitness and immune evasion during the infection process. A multitude of virulence factors and regulators as well as quorum sensing are able to contribute to the versatile interplay between pathogenic bacteria and their hosts. Many of these factors are exposed on the pathogen´s surface. Among these, lipoproteins have attracted considerable attention in recent years as components of ABC transporters and due to their impact on bacterial colonization. However, our knowledge about their roles in pathogen-host interactions and immune evasion is still very limited. In addition, pathogenic bacteria have to adapt to various host niches and hence, regulators are of utmost important in these processes. The impact of lipoproteins on virulence, how these lipoproteins are regulated, and which factors of B. pseudomallei are regulated by QS-sensing systems will be analysed in project area B.
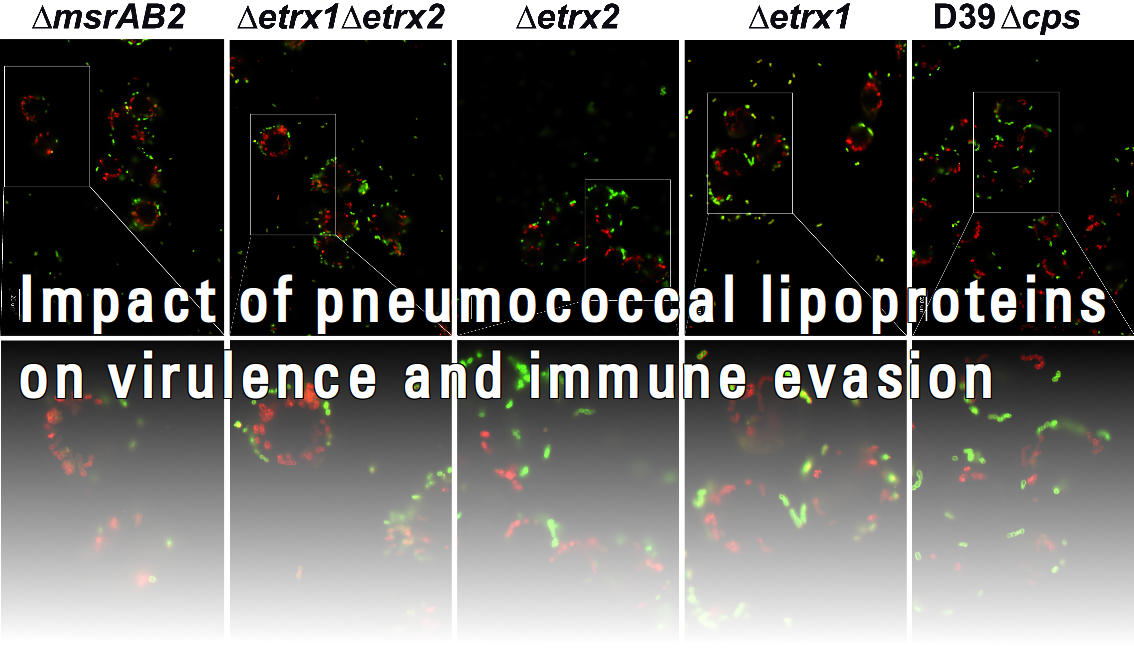
The infectious process of Streptococcus pneumoniae depends on the intracellular toxin pneumolysin and numerous surface-exposed virulence factors. It is widely accepted that a multitude of factors are involved in the initial processes of pneumococcal pathogenesis since depleting individual surface proteins reduced, but did not completely abolish adherence, colonization and invasion of host cells. The cell surface of Streptococcus pneumoniae displays three classical clusters of proteins: choline-binding proteins (CBPs), LPXTG-anchored proteins and lipoproteins. Lipoproteins have recently received considerable attention, since they are known to be essential for the substrate transport due to their function as part of ABC transporter systems, but they are also involved in bacterial fitness and virulence. However, a systematic approach to decipher their role in pathogen-host interactions and immune evasion has not been conducted so far. Loss of function of these proteins was shown to reduce adherence, cause sensitivity to oxidative stress and attenuate virulence in mouse infection models. In this project we will focus on surface-exposed lipoproteins of unknown function and study their impact on pneumococcal-host interactions.
Principal Investigators
Prof. Dr. rer. nat. Sven Hammerschmidt
Thesis Topics
Franziska Voß
"Investigation of the immunogenicity and protective efficacy of surface-localized lipoproteins of Streptococcus pneumoniae"
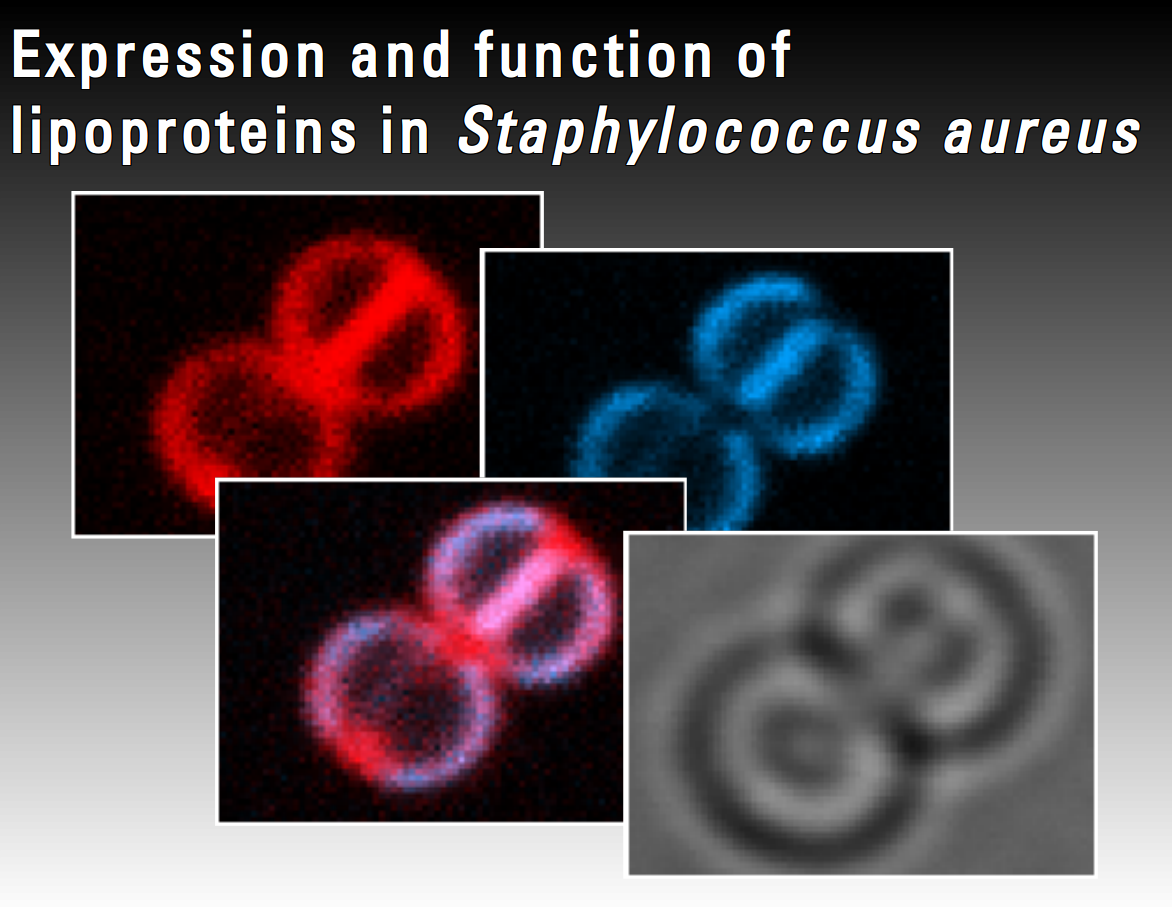
Staphylococcus aureus infections are one of the major threats in hospitals which are often difficult to treat due to the high antibiotic resistance of this pathogen against the common used antibiotics. Thus, there is an urgent need for a better understanding of the pathogenicity of S. aureus. Lipoproteins are retained to the outer membrane cell wall interface via a diacylglyceryl moiety and represent a reservoir of bacterial virulence factors. The present project focuses on the characterization of σB-dependent lipoproteins with so far unknown function. Expression of these proteins will be studied under infection related in vitro conditions using transcriptomic and proteomic techniques. Fluorescence imaging techniques will be performed to analyse in vivo expression and localization of these proteins.
Principal Investigators
Dr. rer. nat. Jan Pané-Farré
Thesis Topics
Anica Beyer
"Expression and Function of Lipoproteins in Staphylococcus aureus"
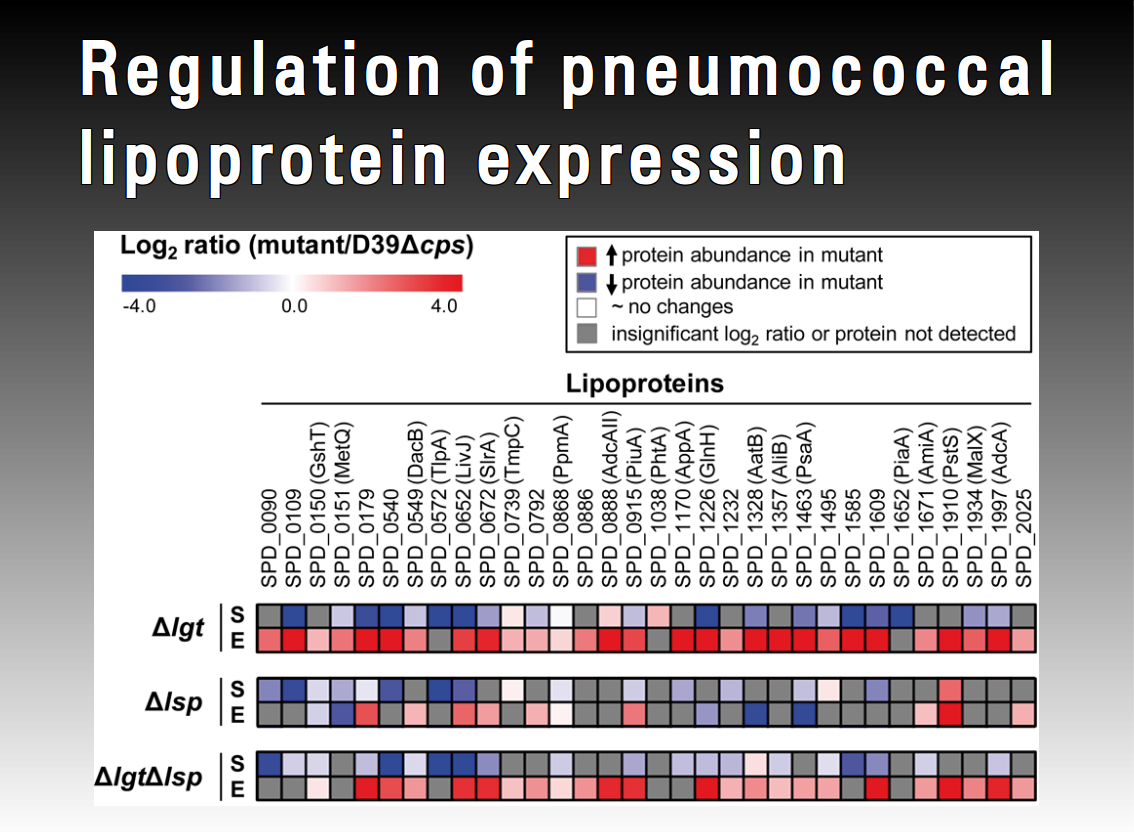
Pneumococcal fitness is of prime importance to survive and multiply during the infectious process. Lipoproteins are a major class of pneumococcal proteins with broad ranging functionality, anchored to the outer layer of the plasma membrane. As extracellular components of ABC transporter systems they greatly affect physiology and bacterial fitness due to their biological function as substrate binding proteins. Loss of function of other lipoproteins influences protein folding, sensitivity to oxidative stress and virulence. The diacylglycerol transferase (Lgt) and lipoprotein signal peptidase (Lsp) are indispensable for the export and maturation of lipoproteins in Gram-positive bacteria such as pneumococci. pneumococcal virulence. This project aims to elucidate regulatory circuits of pneumococcal lipoproteins.
Principal Investigators
Prof. Dr. rer. nat. Sven Hammerschmidt
Thesis Topics
1. Alejandro Gómez Mejia
"Regulation of pneumococcal lipoprotein expression"
2. Jolien Seinen
"Respiratory infections in ICU patients"
Burkholderia pseudomallei employs N-acyl homoserine lactone (AHL) -mediated cell-to-cell communication (“quorum sensing”; QS) to regulate gene expression in a popula-tion density-dependent manner. The genome of B. pseudomallei encodes four four luxI and six luxR homologues. QS-deficient mutants were found to be attenuated in a Syrian hamster pathogenicity model system. Interestingly, some potential virulence factors appear to be down-regulated by QS suggesting that the QS-regulation of pathogenicity may involve so far unknown factors. So far, no systematic analysis of the entire gene/protein expression affected by the different QS-systems of this pathogen has been undertaken. The proposed project aims at systematically characterizing the QS circuitry in B. pseudomallei, since we hypothesize that QS has an important impact on the expression of specific functions related to pathogenicity.
Principal Investigators
Prof. Dr. rer. nat. Katharina Riedel
Thesis Topics
Karolin Stoll-Ziegler
"Impact of quorum-sensing regulated phenotypic traits on the pathogenicity of Burkholderia mallei"
Mucosal surfaces, namely epithelial cells and secreted mucus, constitute a physical barrier that prevents pathogens from gaining access to deeper tissues. In addition, pathogenic bacteria colonizing mucosal surfaces face the innate immune system, including the alternative complement pathway, anti-microbial agents, such as defensins and sentinel cells: professional phagocytes and antigen presenting cells (APCs). Together, they represent the first line immune defence. In parallel, the sentinel cells induce an adaptive immune response resulting in antibody production, T cell effector functions and immune memory. Not surprisingly, pathogens have evolved a multiplicity of strategies to escape host immune responses and persist or disseminate in the organism. The mechanisms of immune recognition of respiratory pathogens, their ingestion and intracellular fate, and conversely, the contribution of specific bacterial components to subvert immune responses are the focus area of the projects in section C.
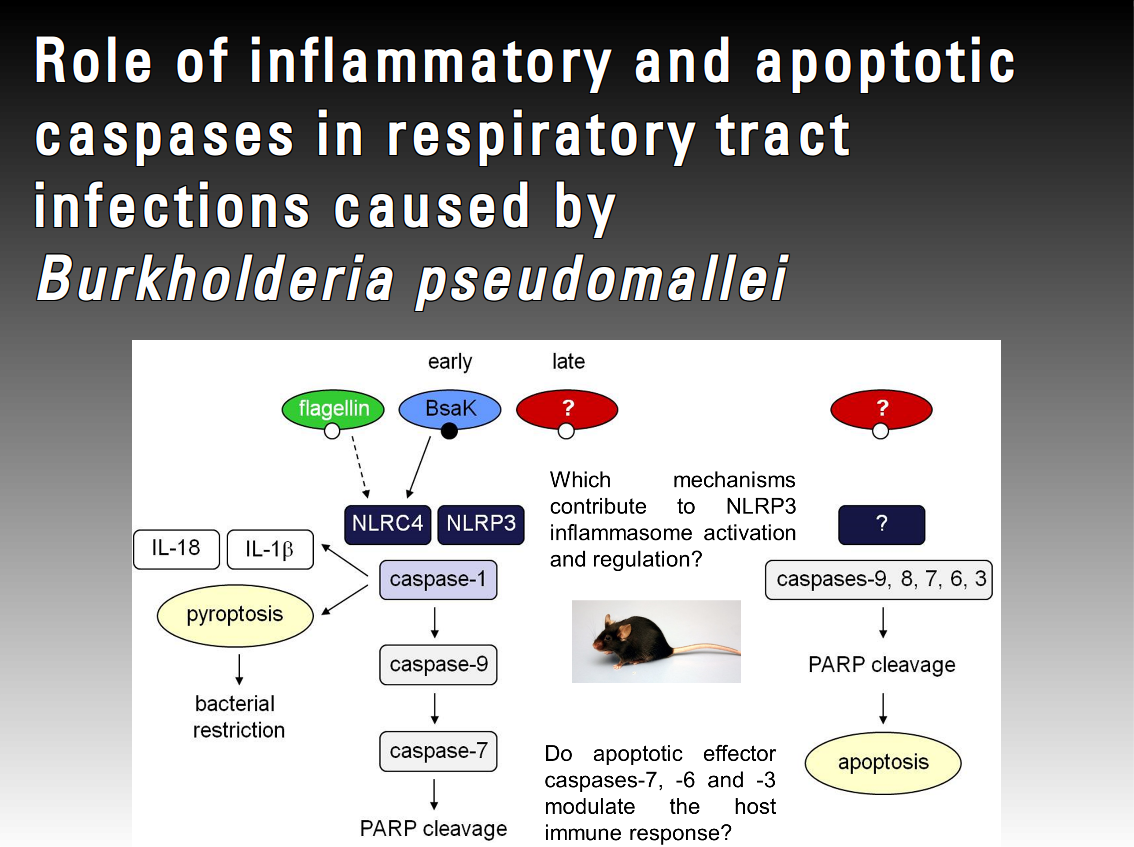
It has been shown for several bacterial pathogens that infection-induced inflammasome assembly leading to caspase-1 activation is a crucial event during early infection. A recent study of Burkholderia pseudomallei lung infection suggests that the Nod-like receptor NLRP3 regulates caspase-1 dependent protective IL-18 and deleterious IL-1β production, whereas NLRC4 regulates caspase-1 dependent pyroptosis, which is important for resistance in this model. However, a possible cross-talk in caspase signalling, in particular caspase-1-dependent activation of apoptotic effector caspases and its possible biological role in pathogenesis of infection has not been addressed so far. This project will focus on caspase-1 activation and caspase-1 mediated effector mechanisms in B. pseudomallei-infected primary macrophages. We want to elucidate the role of small GTPases, ROS production and cathepsin B release in B. pseudomallei-induced NLRP3 inflammasome activation.
Principal Investigators
Prof. Dr. med. Ivo Steinmetz
Thesis Topics
Julia Stiehler
"Role of inflammatory and apoptotic caspases in respiratory tract infections caused by Burkholderia pseudomallei"
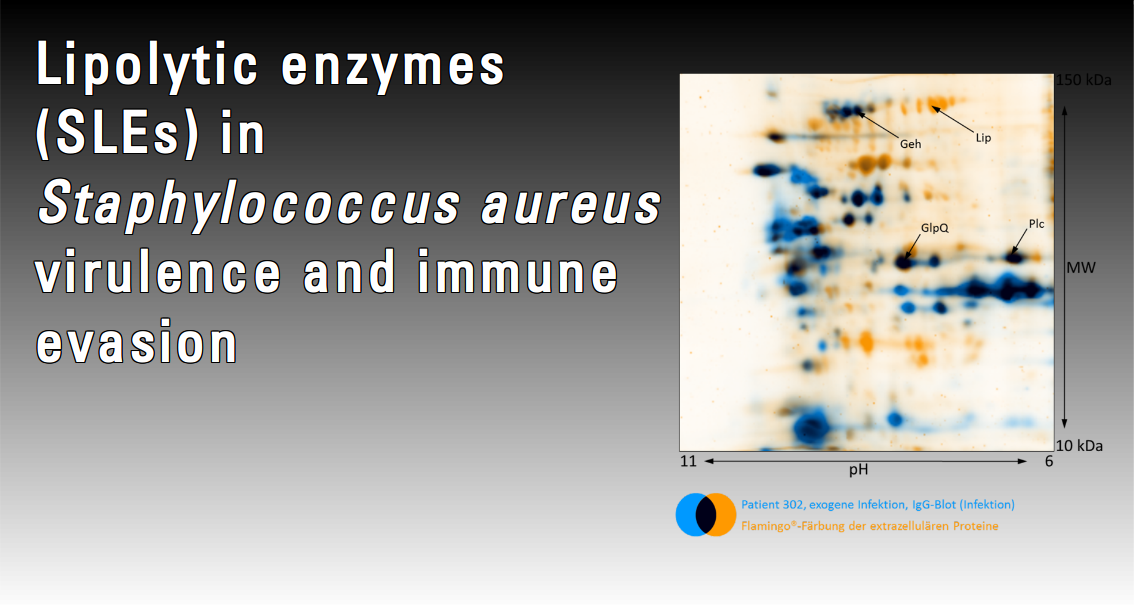
The Staphylococcus aureus genome encodes a number of lipid- and phospholipid-metabolizing enzymes, or lipolytic enzymes (SLEs). These comprise a glycerol ester hydrolase and a lipase (Lip and Geh), a phospholipase (Plc) and two putative glycerophosphoryl diester phophodiesterases (GlpQs), a membrane-bound and a secreted enzyme. All SLEs are abundantly released during the post-exponential growth phase of S. aureusin vitro and they probably contribute to virulence, e.g. by interfering with neutrophil action or by degrading pulmonary surfactant. Intriguingly, the humoral immune response directed against the SLEs differs dramatically: antibodies to Lip are rare, even following bacterial invasion, while antibody titres to its structural homologue Geh are more variable. In contrast, Plc and GlpQ are immunodominant. We propose that SLEs are not only recognized as conventional antigens, but also manipulate the host immune response, thereby contributing to S. aureus virulence and immune evasion.
Principal Investigators
Prof. Dr. med. Barbara M. Bröker
Thesis Topics
Johannes Dick
"Lipase 1 of Staphylococcus aureus and its influence on the immune system"
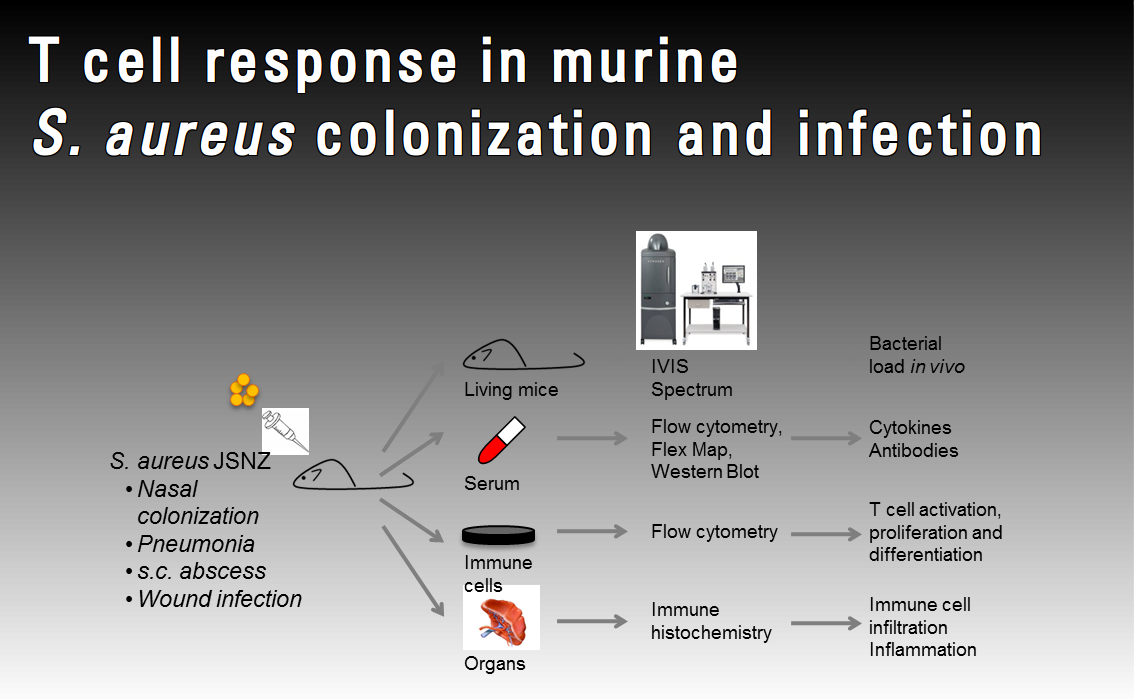
Staphylococcus aureus vaccine development recently opened a new chapter when T cells, in particular Th17 cells, were observed to play a role in S. aureus pathogenesis by enhancing neutrophil recruitment and phagocytosis. However, our knowledge about the T cell response to S. aureus antigens in colonization and infection is still very limited. Using our mouse-adapted model strain JSNZ, we aim to study the T-cell response in murine colonization and infection models and determine how S. aureus manipulates this response. Moreover, we will use OVA as a model antigen to study the antigen-specific T cell response in vivo.
Principal Investigators
Dr. rer. nat. Silva Holtfreter
Thesis Topics
Patricia Trübe
"S. aureus colonization and infection in the mouse model - Tracking T cell responses and their manipulation by bacterial virulence factors"
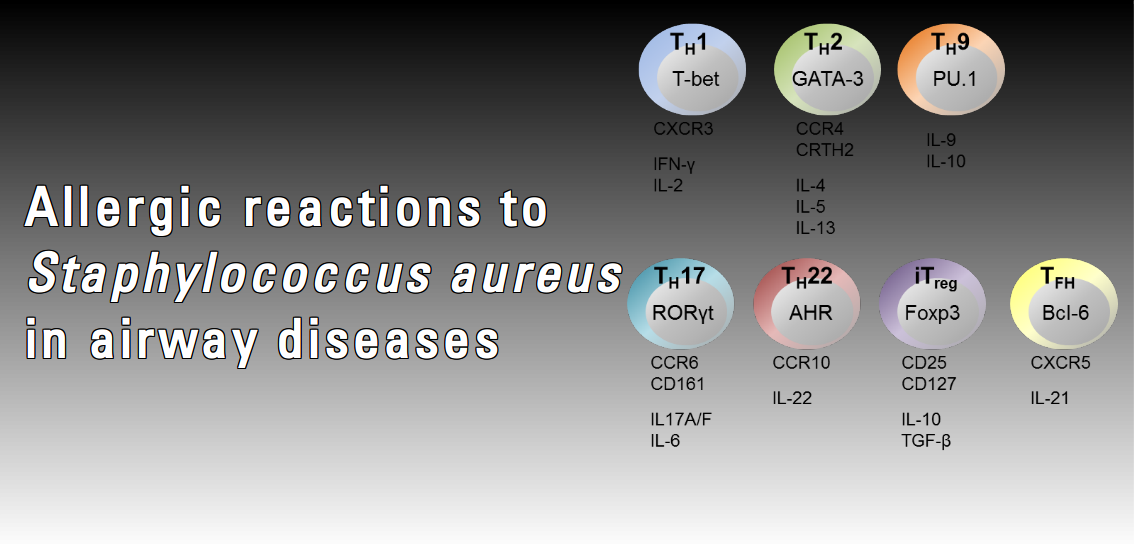
There is increasing evidence that besides commensal and invasive behaviour, S. aureus may also drive allergic reactions. Allergic airway diseases such as rhinosinositis, nasal polyposis and intrinsic asthma are frequently associated with high titre IgE binding to selected staphylococcal superantigens. Superantigens activate large fractions of (memory) T cells, which are thought to exacerbate the allergic inflammation. Intrinsic asthma, a severe respiratory disease largely refractory to treatment, appears to differ from allergic asthma mainly by the absence of sensitization to common allergens. In this disease, a causative role for S. aureus superantigens has been proposed. However, around 20% of clinical S. aureus isolates do not harbour superantigen genes. We hypothesize that in respiratory inflammation superantigens are indicator antigens, i.e. biomarkers of a general allergic reaction to S. aureus. We propose that in susceptible individuals colonization or infection with S. aureus can induce a type 1 hypersensitivity reaction to the bacteria (allergy). We intend to characterize the allergome of S. aureus, the sum of IgE-binding bacterial proteins.
Principal Investigators
Prof. Dr. med. Barbara M. Bröker
Thesis Topics
Maria Nordengrün
"The S. aureus allergome: functional effects of IgG4-binding proteins"